
利用计算生物学、遗传学和微生物学方法,以纤毛虫原生动物、细菌、酵母等为研究模式,研究微生物演化,聚焦生物的突变规律和生活史演化,具体研究主题如下:
A. 土壤原生动物的生物多样性和促进植物生长的机制
B. 土壤原生动物生物肥料研发
C. 原生动物生活史的调控与演化
D. 环境胁迫下的病原微生物的突变与演化
Research interests
We are interested in evolution of prokaryotic and eukaryotic microbes, with a focus on mutation rate and life history evolution. The research models are quite broad, including ciliates, bacteria, yeast, etc. Our research strategy combines population genetics theory, experimental evolution, -omics sequencing, genetic techniques, and bioinformatic tools. The lab deems the training of independent researchers as our first task and research as the second. Graduate students would receive complete training of scientific research and multiple undergrad assistants are also provided for their research. We are now exploring the following topics:
A. Soil protozoa: biodiversity and mechanisms of plant growth promotion
B. Development of soil protozoa-based bio-fertilizers
C. Regulation and evolution of life history in protozoa
D. Mutation and evolution of pathogens under environmental stress
招聘:协助拟加入人员申请中国海洋大学“青年英才工程”(http://rsc.ouc.edu.cn/04/50/c1485a66640/page.htm)等各层次人才计划;定期招收实验室管理、技术等科研助理岗位。
招生:常年招收优秀本科生兼职助理、硕士生、博士生和博士后。
具体信息请邮件咨询(longhongan@ouc.edu.cn)
国家自然科学基金面上项目(2023.01-2026.12;2025.01-2028.12; National Natural Science Foundation of China fundings)
山东省“泰山学者”特聘教授项目(2025.01-2029.12)
国家自然科学基金国际(地区)合作项目(2019.10-2024.9; NSFC-NSF Dimensions 3D collaborative grant)
国家海外高层次人才引进项目(2019-2021)
Recent publications
Pan, J.; Wang, Y.; Li, C.; Zhang, S.; Ye, Z.; Ni, J.; Li, H.; Li, Y.; Yue, H.; Ruan, C.; Zhao, D.; Jiang, Y.; Wu, X.; Shen, X.; Zufall, R.A.; Zhang, Y.; Li, W.; Lynch, M.; Long, H. 2024. Molecular basis of phenotypic plasticity in a marine ciliate. The ISME Journal 18: wrae136 [![]() Pan et al. ISME J 2024.pdf]
Pan et al. ISME J 2024.pdf]
Li, H.; Wu, K.; Feng, Y.; Gao, C.; Wang, Y.; Zhang, Y.; Pan, J.; Shen, X.; Zufall, R.A.; Zhang, Y.; Zhang, W.; Sun, J.; Ye, Z.; Li, W.; Lynch, M.; Long, H. 2024. Integrative analyses on the ciliates Colpoda illuminate the life history evolution of soil microorganisms. mSystems 9: e01379-23 [![]() Li et al. mSystems 2024.pdf]
Li et al. mSystems 2024.pdf]
Zhang, Y.; Li, H.; Wang, Y.; Nie, M.; Zhang, K.; Pan, J.; Zhang, Y.; Ye, Z.; Zufall, R.; Lynch, M.; Long, H. 2024. Mitogenomic architecture and evolution of the soil ciliates Colpoda. mSystems 9: e01161-23 [![]() Zhang et al. mSystems 2024.pdf]
Zhang et al. mSystems 2024.pdf]
Long, H.; Johri, P.; Gout, J.F.; Ni, J.; Hao, Y.; Licknack, T.; Wang, Y.; Pan, J.; Berenice Jiménez-Marín; Lynch, M. 2023. Paramecium genetics, genomics and evolution. Annual Review of Genetics 57: 391-410 [![]() Long et al. ARG 2023.pdf]
Long et al. ARG 2023.pdf]
Lynch, M.; Ali, F.; Lin, T.; Wang, Y.; Ni, J.; Long, H. 2023 The divergence of mutation rates and spectra across the Tree of Life. EMBO Reports e57561 [![]() Lynch et al. EMBO Reports 2023.pdf]
Lynch et al. EMBO Reports 2023.pdf]
Ni, J.; Pan, J.; Wang, Y.; Chen, T.; Feng, X.; Li, Y.; Lin, T.; Lynch, M.; Long, H.; Li, W. 2023. An integrative protocol for one-step PCR amplicon library construction and accurate demultiplexing of pooled sequencing data. Marine Life Science and Technology 5:564-572. [![]() Ni et al. MLST 2023.pdf]
Ni et al. MLST 2023.pdf]
Wu, K.; Li, H.; Cui, X.; Feng, R.; Chen, W.; Jiang, Y.; Tang, C.; Wang, Y.; Shen, X.; Liu, Y.; Lynch, M.; Long, H. 2022. Mutagenesis and resistance development of bacteria challenged by silver nanoparticles. Antimicrobial Agents and Chemotherapy [![]() Wu et al. AAC 2022.pdf]
Wu et al. AAC 2022.pdf]
Pan, J.; Li, W.; Ni, J.; Wu, K.; Konigsberg, I.; Rivera, C.; Tincher, C.; Gregory, C.; Zhou, X.; Doak, T.; Lee, H.; Wang, Y.; Gao, X.; Lynch, M.; Long, H. 2022. Rates of mutations and transcript errors in the foodborne pathogen Salmonella enterica subsp. enterica. Molecular Biology and Evolution 39(4): msac081[![]() Pan et al. MBE 2022.pdf]
Pan et al. MBE 2022.pdf]
Long, H.; Miller, S.F.; Williams, E.; Lynch, M. 2018. Specificity of the DNA mismatch repair system (MMR) and mutagenesis bias in bacteria. Molecular Biology and Evolution 35(10):2414-2421. [![]() Long et al Mol Biol Evol 2018.pdf]
Long et al Mol Biol Evol 2018.pdf]
Long, H.; Sung, W.; Kucukyildirim,S.; Williams,E.; Miller, S.;Guo, W.; Patterson, C.; Gregory, C.; Strauss, C.; Stone, C.; Berne, C.; Kysela, D.; Shoemaker, W.; Muscarella, M.; Luo, H.; Lennon, J.; Brun, Y.; Lynch, M. 2018. Evolutionary determinants of genome-wide nucleotide composition. Nature Ecology and Evolution 2: 237-240. [![]() Long et al Nat Ecol Evol 2018.pdf]
Long et al Nat Ecol Evol 2018.pdf]
Long, H.; Miller, S.F.; Strauss, C.; Zhao, C.; Cheng, L.; Ye, Z.; Griffin, K.; Te, R.; Lee, H.; Chen, C.; Lynch, M. 2016. Antibiotic treatment enhances the genome-wide mutation rate of target cells. PNAS 113(18): E2498-E2505. [![]() Long et al. PNAS 2016.pdf]
Long et al. PNAS 2016.pdf]
Long, H.; Kucukyildirim#, S.; Sung, W.; Williams, E.; Lee, H.; Ackerman, M.; Doak, T.; Tang, H.; Lynch, M. 2015. Background mutational features of the radiation-resistant bacterium Deinococcus radiodurans. Molecular Biology and Evolution 32(9): 2383-2392. [![]() Long et al. MBE 2015.pdf]
Long et al. MBE 2015.pdf]
2024 山东省科普创作大赛一等奖
2021 中国海洋大学优秀教师
2019 中国海洋大学第21届天泰奖一等奖;中国海洋大学优秀班主任;中国海洋大学第20届五四青年奖
2018 李氏基金会杰出创新奖
2016 美国遗传学会DeLill Nasser遗传学职业发展奖
2018 Li Foundation Heritage Prize-Excellence in Creativity
2016 DeLill Nasser Award for Professional Development in Genetics, Genetics Society of America

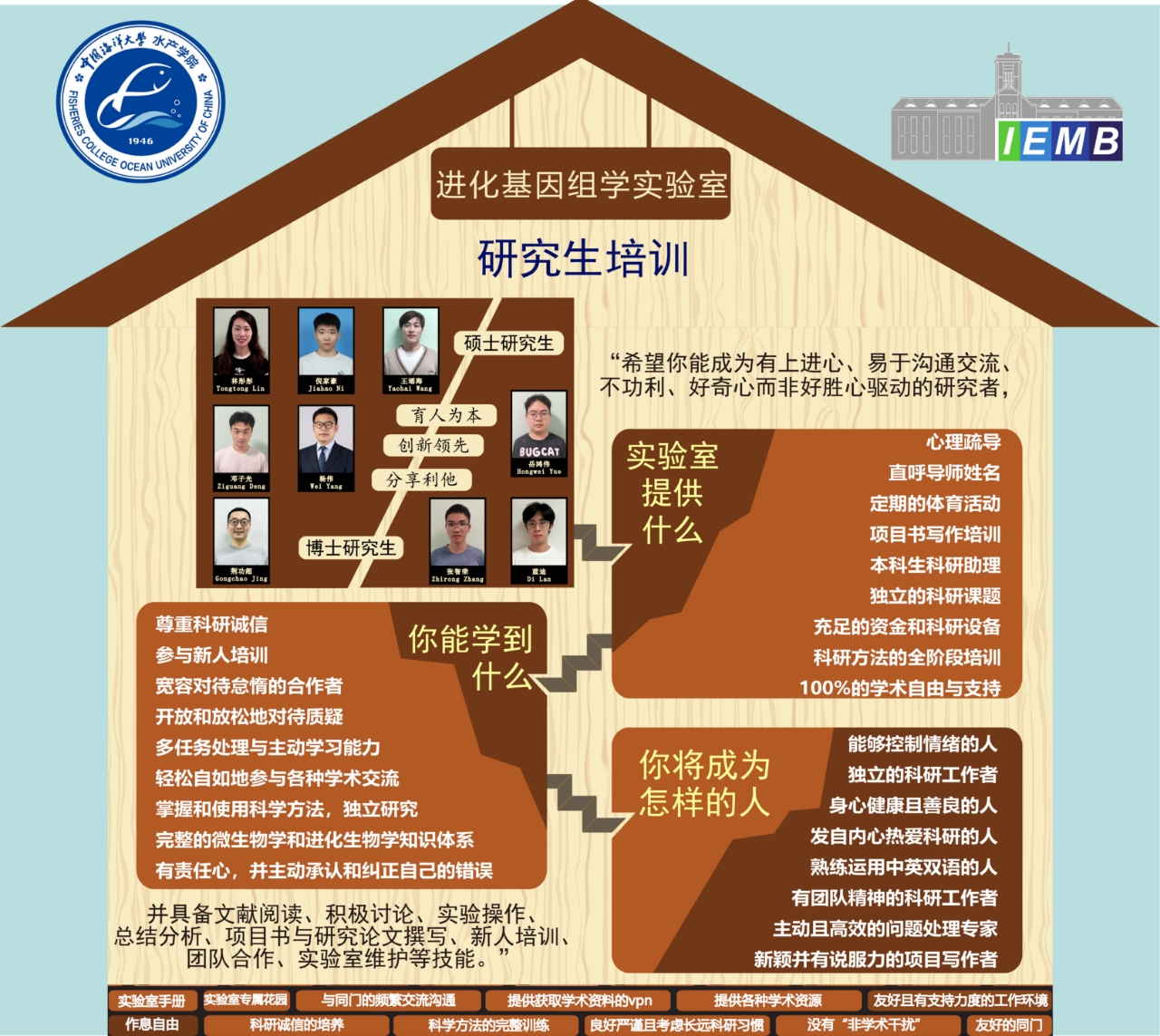
育人为本,创新领先,分享利他

博士研究生:
林彤彤(2021博士研究生),水生生物学,lintongtong@stu.ouc.edu.cn,微生物基因组的极端演化
倪家豪(2022博士研究生),水生生物学,nijiahao@stu.ouc.edu.cn,纤毛虫多样性的产生与维持
蒋雨柔(2022博士研究生),水生生物学,jiangyurou@stu.ouc.edu.cn,微生物演化模型构建
舒王欣泽(2022博士研究生),水生生物学,11220511010@stu.ouc.edu.cn,环境微生物学
王瑶海(2023博士研究生),水生生物学,wangyaohai@stu.ouc.edu.cn,纤毛虫逆境生存机制
邓子光(2024博士研究生),水生生物学,dengziguang@stu.ouc.edu.cn,土壤纤毛虫生物多样性与演化
荆功超(2024博士研究生,在职),生物与医药,11240523005@stu.ouc.edu.cn,肠道沙门氏菌的演化
杨伟(2024博士研究生,在职),生物与医药,yangwei@ouc.edu.cn,肺炎克莱伯氏菌的演化
硕士研究生:
岳鸿伟(2024硕士研究生),水产养殖学,yuehongwei@stu.ouc.edu.cn,土壤纤毛虫生物多样性与演化
张智荣(2024硕士研究生),水生生物学,zhangzhirong@stu.ouc.edu.cn,病原菌的突变规律
蓝迪(2024硕士研究生),水产养殖学,landi@stu.ouc.edu.cn,环境污染物影响下的细菌突变规律与耐药机制
已毕业:
博士:潘娇 2023.06 (PHD 001),吴琨 2023.06 (PHD 002),李海潮 2024.06 (PHD 003),蒋婉月 2024.06 (PHD 004)
硕士:周霞 2023.06(MS 001),张渊渊 2024.06 (MS 002)
共享技术
1. 一步PCR超快扩增子文库构建(OSPALC):![]() Ni et al. MLST 2023.pdf, 中文介绍(
Ni et al. MLST 2023.pdf, 中文介绍(![]() OSPALC一步扩增子文库构建法.pdf),实验手册(
OSPALC一步扩增子文库构建法.pdf),实验手册(![]() OSPALC扩增子文库构建手册-中文版(1).pdf)
OSPALC扩增子文库构建手册-中文版(1).pdf)
2. 基因组文库构建缩量法(Li et al. MLST 2019)
3. 磁珠筛选用磁力架,3D模型(Li et al. MLST 2019, File S2-S5)
4. 《生物大数据分析》操作培训:新闻条介绍
教育题材小说《转变 第一部》
(ISBN号:978-7-5670-2188-4,出版中):
链接:https://pan.baidu.com/s/1rnDdB3HSX_Q3Vw649Dw2Qw
提取码:2rge
原生动物主题科普绘本“微虫小记”丛书
(ISBN号:978-7-5670-3308-5,2022年11月出版,中国海洋大学出版社,主编:龙红岸):
实验室勤工助学淘宝网页:https://m.tb.cn/h.gML9EjWfUzF4N9M?tk=94i63gZKZeKMF6563
Free lab strains upon request: Chaenea PJ13002; Uronema PJ20101A; Glauconema LHA0827; Glauconema LJL43
Please contact Dr. Hongan Long: longhongan@ouc.edu.cn