
空格
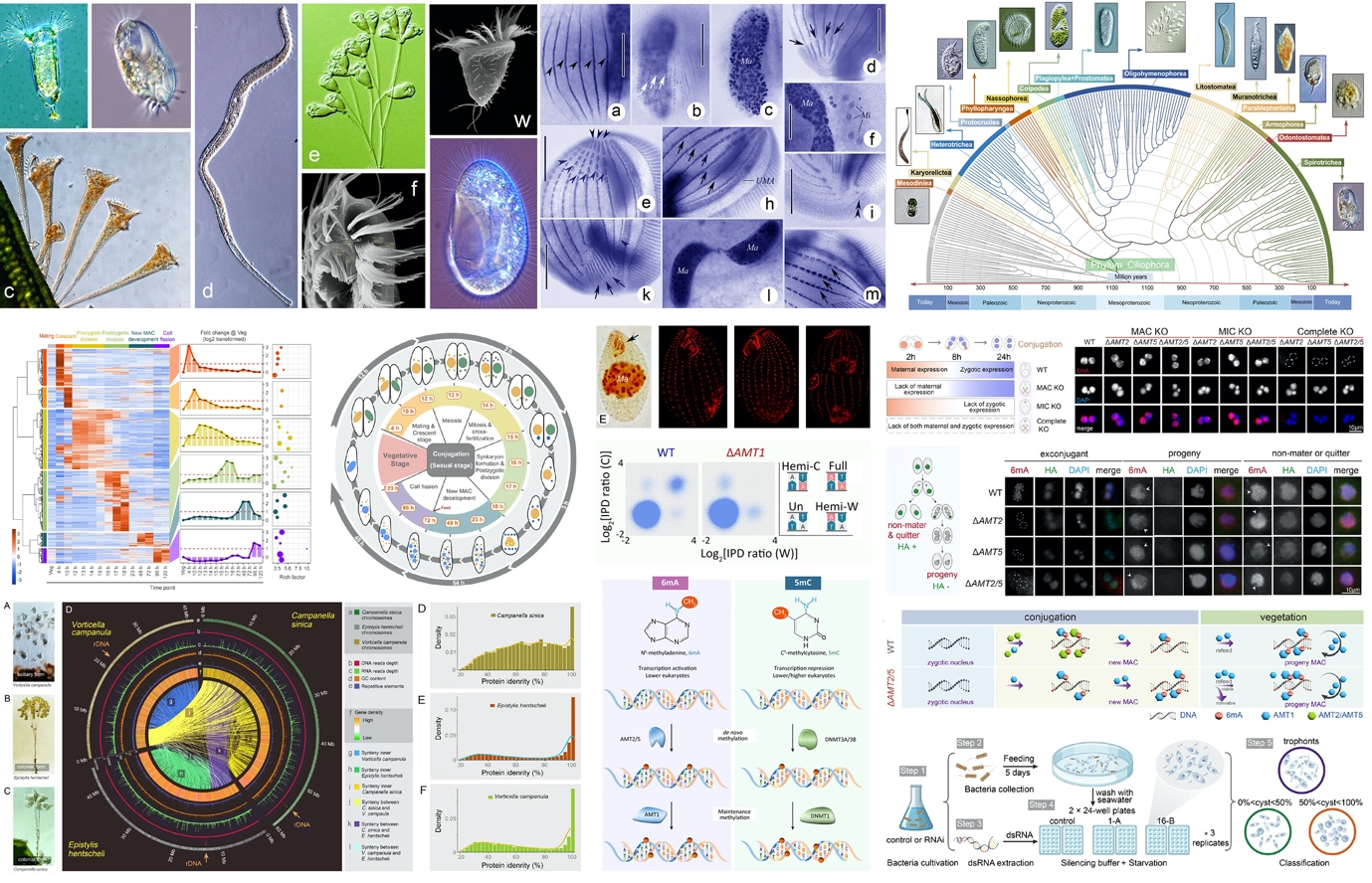
空格围绕纤毛虫原生动物的多样性与系统演化、细胞结构的发生与分化、表观遗传调控、性别决定、基因组重排、基因进化等基础科学问题,主要研究包括:
(1)聚焦我国温带、亚热带-热带沿海与内陆湿地环境中的自由生活类群,研究其区系、形态分类、物种多样性、群落结构、生态学功能与地位、时空与地理分布;
(2)以四膜虫为材料,探讨以甲基化修饰为主的表观遗传信息对基因表达和复制的调控机制;
(3)以游仆虫、草履虫等为材料,探讨纤毛虫有性生殖过程中大、小核发育、分化及基因组重排的模式和机制,解析其性别分化和决定的相关基因及分子机制;
(4)围绕纤毛虫的系统演化、分化过程中的重要事件和组学特征,探讨细胞及胞器分化、适应与进化、结构与功能关系等重要现象和过程中相关功能基因的调控机制;
(5)以草履虫为材料,聚焦胞质分裂调控的分子机制,探究收缩环的形成、细胞极性的建立和维持;
(6)探讨纤毛虫无性及有性生殖期间细胞器结构及模式的形成、分化等个体发育特征;
(7)构建纤毛虫各代表类群的DNA库、标记基因库、组学信息库等遗传信息库; 探讨纤毛门内各高阶阶元间的亲缘关系、系统发育与演化。
空Focusing on fundamental research related to ciliated protozoa, including biodiversity and phylogeny, morphogenesis and differentiation of subcellular structures, epigenetic regulation, sex determination, genomics and genome rearrangement, gene evolution, etc. Our current focuses are:
空(1) Taxonomy and biodiversity. Focusing on free-living ciliates in coastal and inland wetlands across temperate and subtropical-tropical regions in China, explore their fauna, morphological classification, species diversity, community structure, ecological functions, and spatiotemporal and geographical distributions.
空(2) Epigenetics. Understand how DNA and histone methylations affect gene expression and DNA replication by using the model ciliate Tetrahymena thermophila.
空(3) Genome rearrangement and sex determination. Investigate the patterns and mechanisms of nucleus development, differentiation, and genome rearrangements during sexual reproduction in representative ciliates, and explore the evolution and molecular mechanisms of sex determination, using Euplotes and Paramecium as model organisms.
空(4) Genome evolution. Explore the regulatory mechanisms of functional genes involved in key phenomena and processes of ciliates, such as cell and organelle differentiation, speciation and adaptation, applying combined multi-omics methods.
空(5) Cytokinesis. Using Paramecium as model organism, focus on the molecular mechanism of cytokinesis regulation, including the contractile ring formation, and the establishment and maintenance of cell polarity.
空(6) Cell development. Focus on the formation, differentiation, and developmental characteristics of organelle structures and patterns during sexual and asexual reproduction in ciliates.
空(7) Phylogenetics and systematics. Investigate the phylogeny, systematics, and evolutionary relationships among diverse lineages within the phylum Ciliophora, while constructing genetic information databases containing genomic DNA, marker gene sequences and omics data for representative ciliate groups.
· 国家自然科学基金“杰青”项目:纤毛虫原生动物基础生物学(2022-2026)
· 国家自然科学基金重点项目:淡水湿地中纤毛虫原生动物多样性格局与档案资料的建立:以东湖(武汉)与微山湖水系为例(2021-2025)
· 国家自然科学基金面上项目:尾草履虫有性生殖过程中大规模DNA删除的分子机制探究(2025-2028)
· 国家自然科学基金面上项目:多小核草履虫交配型昼夜节律逆转的分子机制探究(2023-2026)
· 国家自然科学基金面上项目:以海洋纤毛虫为材料:大小核基因重组模式及机制探讨(2023-2026)
· 国家自然科学基金青年项目:以嗜热四膜虫为模式:真核生物DNA N6-腺嘌呤甲基化的关键调控因子的作用机制研究(2023-2025)
·国家自然科学基金青年学生基础研究项目:微山湖湿地中的侧口目纤毛虫:多样性与分子资源库构建(2025-2026)
· 山东省自然科学基金重大基础研究项目:以四膜虫为模式:DNA N6-甲基腺嘌呤(6mA)从头甲基化的建立机制和生物学功能研究(2025-2027)
· 山东省“泰山学者”青年专家计划:纤毛虫原生动物的基因进化(2025-2027)
· 山东省“泰山学者”青年专家计划:纤毛虫功能基因及组学研究(2021-2025)
2025 Cheng T., Zhang J., Li H., Diao J., Zhang W., Niu J., Kawaguchi T., Nakayama J., Kataoka K., Gao S. Identification and characterization of the de novo methyltransferases for eukaryotic N6-methyladenine (6mA). Science Advances. 11, eadq4623.
2025 Wang Y.Y., Nan B., Ye F., Zhang Z., Yang W.T., Pan B., Wei F., Duan L.L., Li H.C., Niu J.H., Ju A.L., Liu Y.Q., Wang D.T., Zhang W.X., Liu Y.F., Gao S. Dual modes of DNA N6-methyladenine maintenance by distinct methyltransferase complexes. PNAS, 122(3): e2413037121.
2025 Ye F., Chen X., Li Y.Q., Ju A.L., Sheng Y.L., Duan L.L., Zhang J.C., Zhang Z., Al-Rasheid K.A.S., Stover N.A., Gao S. Comprehensive genome annotation of the model ciliate Tetrahymena thermophila by in-depth epigenetic and transcriptomic profiling. Nucleic Acids Research, gkae1177.
2025 Duan L.L., Li H.C., Ju A.L., Zhang Z., Niu J.H., Zhang Y.M., Diao J.H., Liu Y.Q., Ma H.G., Song N., Kataoka K., Gao S., Wang Y.Y. Methyl-dependent auto-regulation of the DNA N6-adenine methyltransferase AMT1 in the unicellular eukaryote Tetrahymena thermophila. Nucleic Acids Research, gkaf022.
2025 Li H. C., Niu J.H., Sheng Y.L., Liu Y.F., Gao S. SMAC: identifying DNA N6-methyladenine (6mA) at the single-molecule level using SMRT CCS data. Briefings in Bioinformatics, 26(2), bbaf153
2025 Jiang Y.H., Chen X., Wang C.D., Lyu L.P., Al-Farraj S.A., Stover N.A., Gao F. Genes and proteins expressed at different life cycle stages in the model protist Euplotes vannus revealed by both transcriptomic and proteomic approaches. Science China Life Sciences, 68, 232–248.
2025 Zheng W., Li C., Zhou Z.R., Chen X., Lynch M., Yan Y. Unveiling an ancient whole-genome duplication event in Stentor, the model unicellular eukaryotes. Science China Life Sciences, 68, 825–835.
2024 Wang C.D., Lyu L.P., Solberg T., Zhang H.Y., Wen Z., Gao F. GTSF1 is required for transposon silencing in the unicellular eukaryote Paramecium tetraurelia. Nucleic Acids Research,52: 13206–13223.
2024 Liu Y.Q., Niu J.H., Ye F., Solberg T., Lu B.R., Wang C.D., Nowacki M., Gao S. Dynamic DNA N6-adenine methylation (6mA) governs the encystment process, showcased in the unicellular eukaryote Pseudocohnilembus persalinus. Genome Research, 34(2): 256−271.
2024 Sheng Y.L., Wang Y.Y., Yang W.T., Wang X.Q., Lu J.W., Pan B., Nan B., Liu Y.Q., Ye F., Li C., Song J.K., Dou Y.L., Gao S., Liu Y.F. Semiconservative transmission of DNA N6-adenine methylation in a unicellular eukaryote. Genome Research, 34(5): 740−756.
2024 Gao Y.Y., Solberg T., Wang R., Yu Y.E., Al-Rasheid K., Gao F. Application of RNA interference and protein localization to investigate housekeeping and developmentally regulated genes in the emerging model protozoan Paramecium caudatum. Communications Biology, 7, 204.
2024 Li T., Zhang T., Liu M., Zhang Z., Zhang J., Niu J., Chen X., Al Farraj S.A., Song W.B. Findings on three endocommensal scuticociliates (Protista, Ciliophora) from freshwater mollusks, including their morphology and molecular phylogeny with descriptions of two new species. Marine Life Science & Technology, 6: 212-235.
2023 Pan B., Ye F., Li T., Wei F., Warren A., Wang Y.Y., Gao S. Potential role of N6-adenine DNA methylation in alternative splicing and endosymbiosis in Paramecium bursaria. iScience, 26(5): 106676.
2023 Gao Y.Y., Solberg T., Wang, C.D., Gao F. Small RNA-mediated genome rearrangement pathways in ciliates. Trends in Genetics, 39(2): 94−97.
2023 Lu B.R., Hu X.Z., Warren A., Song W.B., Yan Y. From oral structure to molecular evidence: new insights into the evolutionary phylogeny of the ciliate order Sessilida (Protista, Ciliophora), with the establishment of two new families and new contributions to the poorly studied family Vaginicolidae. Science China Life Sciences, 66: 1535–1553.
2022 Wang C.D., Solberg T., Maurer-Alcalá X.X., Swart C.E., Gao F., Nowacki M. A small RNA guided PRC2 complex eliminates DNA as an extreme form of transposon silencing. Cell Reports, 40: 111263.
2022 Qiao Y., Cheng T., Zhang J., Alfarraj S.A., Tian M., Liu Y. & Gao S. Identification and utilization of a mutated 60S ribosomal subunit coding gene as an effective and cost-efficient selection marker for Tetrahymena genetic manipulation. International Journal of Biological Macromolecules, 204: 1−8.
2022 Ma M.Z., Xu Y., Lu B.R., Li Y.Q., Song W.B., Petroni G., Al-Farrah SA., & Yan Y. Molecular Phylogeny and taxonomy of four Remanella species (Protozoa, Ciliophora): a flagship genus of karyorelictean ciliates, with descriptions of three new species. Zoological Research, 43: 827−842.
2022 Ma M.Z., Li Y.Q., Maurer-Alcala X., Wang Y.R. & Yan Y. Deciphering phylogenetic relationships in class Karyorelictea (Protista, Ciliophora) based on updated multi-gene information with establishment of a new order Wilbertomorphida n. ord. Molecular Phylogenetics and Evolution, 169: 107406.
2022 Zhang G.A.T., Zhao Y., Chi Y., Warren A., Pan H.B. & Song, W.B. Updating the phylogeny and taxonomy of pleurostomatid ciliates (Protista: Ciliophora) with establishment of a new family, a new genus and two new species. Zoological Journal of the Linnean Society, 196: 105–123.
2021 Xu J., Zhao X.L., Mao F.B., Basrur V., Ueberheide B., Chait B.T., Allis C.D., Taverna S.D., Gao S., Wang W. & Liu Y. A Polycomb repressive complex is required for RNAi-mediated heterochromatin formation and dynamic distribution of nuclear bodies. Nucleic Acids Research, 49, 5407–5425.
2021 Zheng W.B., Wang C.D., Lynch M. & Gao S. The compact macronuclear genome of the ciliate Halteria grandinella: a transcriptome-like genome with 23,000 nanochromosomes. mBio, 12: e01964-20.
2021 Chi Y., Chen X., Li Y., Wang C., Zhang T., Ayoub A., Warren A., Song W. & Wang Y.Y. New contributions to the phylogeny of the ciliate class Heterotrichea (Protista, Ciliophora): analyses at family-genus level and new evolutionary hypotheses. Science China Life Sciences, 64: 606–620.
2021 Liu Y., Nan B., Niu J., Kapler G. & Gao S. An optimized and versatile counter-flow centrifugal elutriation workflow to obtain synchronized eukaryotic cells. Frontiers in Cell and Developmental Biology, 9: 664418.
2021 Wang, C.D., Gao, Y.Y., Lu, B.R., Chi, Y., Zhang, T.T., El-Serehy, H. A., Al-Farraj, S. A., Li, L.F., Song, W.B. & Gao, F. Large-scale phylogenomic analysis provides new insights into the phylogeny of the class Oligohymenophorea (Protista, Ciliophora) with establishment of a new subclass Urocentria nov. subcl. Molecular Phylogenetics and Evolution, 159: 107112.
2021 Zhang T.T., Li C., Zhang X., Wang C.D., Roger A.J. & Gao F. Characterization and comparative analysis of mitochondrial genomes in single-celled eukaryotes shed light on the diversity and evolution of the linear molecular architecture. International Journal of Molecular Science, 22: 2546.
2021 Ma M.Z., Xu Y., Yan Y., Li Y.Q., Warren A. & Song W.B. Taxonomy and molecular information of four karyorelictids in genera Apotrachelocerca and Tracheloraphis (Protozoa, Ciliophora), with notes on their systematic positions and description of two new species. Zoological Journal of the Linnean Society, 192: 690–709.
2021 Sheng Y., Pan B., Wei F., Wang Y.Y. & Gao S. Case study of the response of N6-methyladenine DNA modification to environmental stressors in the unicellular eukaryote Tetrahymena thermophila. mSphere, 6: e01208-20.
2020 Sheng Y., Duan L., Cheng T., Qiao Y., Stover N.A., Gao S. The completed macronuclear genome of a model ciliate Tetrahymena thermophila and its application in genome scrambling and copy number analyses. Science China Life Sciences, 63: 1534–1542.
2020 Zheng W., Chen J., Doak T.C., Song W.B., Yan Y. ADFinder: accurate detection of programmed DNA elimination using NGS high-throughput sequencing data. Bioinformatics, 36: 3632–3636.
2020 Chi Y., Duan L.L., Luo X.T., Cheng T., Warren A., Huang J. & Chen X.R. A new contribution to the taxonomy and molecular phylogeny of three freshwater, well-known species of the ciliate genus Spirostomum (Protozoa: Ciliophora: Heterotrichea). Zoological Journal of the Linnean Society, 189:158–177.
原生动物学研究室网址:http://scxy.ouc.edu.cn/lplb/