
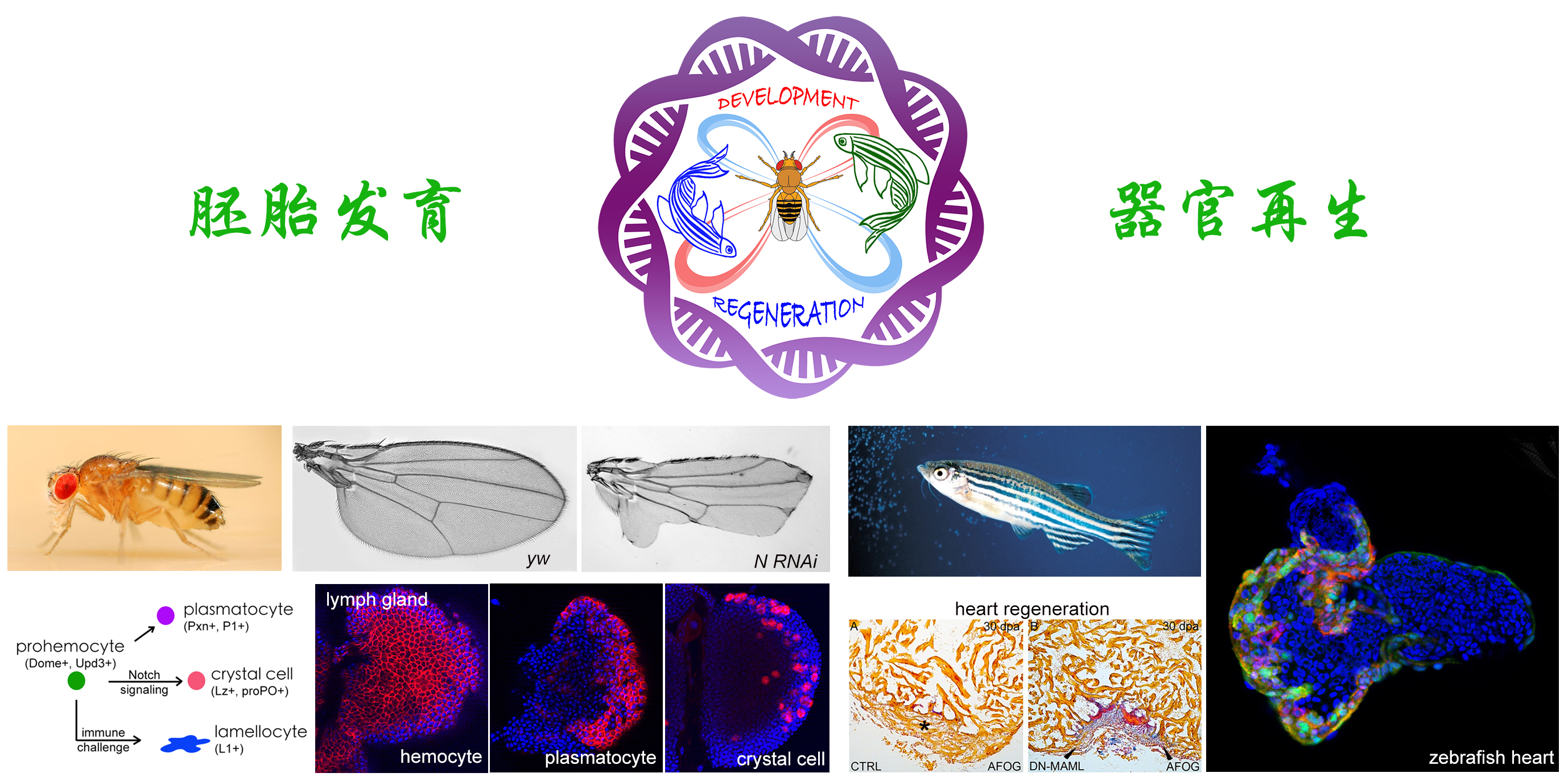
动物胚胎发育和成体器官再生的过程包含了“细胞命运的决定”以及“相关形态结构构建”两个相对独立却相互影响的过程。细胞信号转导是细胞命运决定过程中关键的调控机制:靶细胞通过受体接收信号细胞分泌出来的配体分子,引发其胞内的信号级联反应,最终诱导该细胞向特定命运分化。在细胞命运决定的同时,细胞通过改变其行为,驱动组织层面上的变化,逐步形成预定的结构。
本实验室以果蝇和斑马鱼为主要模式生物,结合遗传学、组学、分子生物学、显微成像与定量分析等现代技术,系统研究Hedgehog、Notch、JNK等多种重要发育信号的相互交织决定细胞命运的分子机理,以及细胞命运决定之后的细胞行为驱动胚胎发育以及器官(心脏)再生的作用机理。
Animal embryonic development and organ regeneration are driven by the signaling network-controlled cell fate determination and cell-shape-based tissue morphogenesis. The target cells receive ligand molecules secreted by the signaling cell, which triggers an intracellular signaling cascade that ultimately induces the cell to differentiate toward a specific fate. While the cell's fate is being determined, interactions between cell-cell contacts, remodeling of the cytoskeleton, together with the cell number control system, collectively define tissue morphology, gradually forming the intended structure.
Using Drosophila and zebrafish as the main model organisms, our laboratory focuses on how the interwoven signaling pathways, such as Hedgehog, Notch, and JNK, control embryo development and organ (mainly heart) regeneration in terms of cell fate determination, and how cellular behavior drives tissue morphogenesis.
·中国海洋大学“筑峰人才工程”启动经费
·中国海洋大学“青年英才工程”启动经费2项
·山东省泰山学者“青年专家”经费
·国家自然科学基金面上项目5项,青年项目1项
本科生课程:
《细胞生物学》理论(40课时,2.5学分,春季学期,水产学院;赵龙,独立讲授)
《分子生物学》理论(双语,48课时,3学分,春季学期,海德学院;苏颖,与其他两位老师合讲)
《结构生物化学》理论(双语,32课时,2学分,秋季学期,海德学院;吕志一,独立讲授)
《模式生物发育与进化》理论(32课时,2.5学分,春季学期,全校通识课;苏颖、赵龙,参与讲授)
研究生课程:
《发育分子生物学》,苏颖、赵龙,参与讲授
《高级生物化学》,赵龙,参与讲授
《学术论文写作》与《论文写作指导》,图像处理部分,吕志一,讲授。
发表论文
*通讯作者,#共同一作
1. Luo W, Liu S, Zhang F, Zhao L*, Su Y*. (2022) Metabolic strategy of macrophages under homeostasis or immune stress in Drosophila. Marine Life Science & Technology 4(3): 291-302.
2. Fu Y#, Lv Z#, Kong D, Fan Y, Dong B*. (2022) High abundance of CDC45 inhibits cellproliferation through elevation of HSPA6. Cell Proliferation 55(7): e13257.
3. Lv Z, Zhang N, Zhang X, Großhans J, Kong D*. (2022) The lateral epidermis actively counteracts pulling by the amnioserosa during dorsal closure. Frontiers in Cell and Developmental Biology 10:865397.
4. Liu M#, Su Y#, Peng J, Zhu AJ. (2021) Protein modifications in Hedgehog signaling: Cross talk and feedback regulation confer divergent Hedgehog signaling activity. Bioessays Nov 5: e2100153.
5. Gan T, Fan L, Zhao L, Misra M, Liu M, Zhang M*, Su Y*.(2021) JNK signaling in Drosophila aging and longevity. International Journal of Molecular Sciences 22(17): 9649.
6. Zhao L #, Gao F #, Gao S #, Liang Y #, Long H #, Lv Z #, Su Y#, Ye N #, Zhang L #, Zhao C #, Wang X, Song W, Zhang S, Dong B. (2021) Biodiversity-based development and evolution: The emerging research systems in model and non-model organisms. Science China Life Sciences64(8): 1236-1280.
7. Gao J, Fan L, Zhao L*, Su Y*. (2021) The interaction of Notch and Wnt signaling pathways in vertebrate regeneration. Cell Regeneration 10(1): 11.
8. Liu M, Liu A, Wang J, Zhang Y, Li Y, Su Y*, Zhu AJ*. (2021) Competition between two phosphatases fine-tunes Hedgehog signaling. Journal of Cell Biology 220(2): e202010078.
9. Schmidt A#, Li L#, Lv Z, Yan S and Großhans J*. (2021) Dia- and Rok-dependent enrichment of capping proteins in a cortical region. Journal of Cell Science 134(21): jcs258973.
10. Lv Z, de-Carvalho J, Telley I and Großhans J*. (2021) Cytoskeletal mechanics and dynamics in the Drosophila syncytial embryo. Journal of Cell Science 134(4). 5.285
11. Lan W, Liu S, Zhao L*, Su Y*. (2020) Regulation of Drosophila hematopoiesis in lymph gland: from a developmental signaling point of view. International Journal of Molecular Sciences 21: 5246.
12. 赵又佼,赵龙*,苏颖* (2020) 锌离子、锌转运蛋白-细胞信号通路的新调控因子。中国细胞生物学学报42(9): 1631-1641.
13. Lv Z*, Rosenbaum J, S Mohr S, Zhang X, Kong D, Preiss H, Kruss S, Alim K, Aspelmeier T and Großhans J*. (2020) The Emergent Yo-yo Movement of Nuclei Driven by Cytoskeletal Remodeling in Pseudo-synchronous Mitotic Cycles. Current Biology 30(13): 2564-2573 e2565.
14. Lu Q, Gao Y, Fu Y, Peng H, Shi W, Li B, Lv Z, Feng X and Dong B*. (2020) Ciona embryonic tail bending is driven by asymmetrical notochord contractility and coordinated by epithelial proliferation. Development 147(24): dev185868.
15. Selvaggio G, Chizhik A, Nissler R, Kuhlemann L, Meyer D, Vuong L, Preiss H, Herrmann H, Mann F, Lv Z, Oswald T, Spreinat A, Erpenbeck L, Großhans J, Karius V, Janshoff A, Giraldo J and Kruss S*. (2020) Exfoliated near infrared fluorescent silicate nanosheets for (bio)photonics. Nature Communications 11(1): 1495.
16. Zhao L, Ben-Yair R, Burns CE, Burns CG. (2019) Endocardial Notch signaling promotes cardiomyocyte proliferation in the regenerating zebrafish heart through Wnt pathway antagonism. Cell Reports 26(3): 546-554.
17. Galvez-Santisteban M, Chen D, Zhang R, Serrano R, Nguyen C, Zhao L, Nerb L, Masutani EM, Vermot J, Burns CG, Burns CE, del Alamo JC, Chi NC. (2019) Hemodynamic-mediated endocardial signaling controls in vivo myocardial reprogramming. eLife 8:e44816.
18. Wang L, Yang J*, Wang H, Ran C, Su Y, Zhao L*. (2019) A highly selective turn-on fluorescent probe detection of aluminum and its application to bio-imaging. Sensors 19(11): 2423.
19. Kong D*, Lv Z, Haring M, Lin B, Wolf F and Großhans J. (2019) In vivo optochemical control of cell contractility at single-cell resolution. EMBO Reports 20(12): e47755.
20. Lv Z, Lu Q, and Dong B*. (2019) Morphogenesis: a focus on marine invertebrates. Marine Life Science & Technology 1(1): 28-40.
21. Mann FA, Lv Z, Großhans J, Opazo F and Kruss S*. (2019) Nanobody-Conjugated Nanotubes for Targeted Near-Infrared In Vivo Imaging and Sensing. Angewandte Chemie International Edition 58(33): 11469-11473.
22. Chi C, Wang L, Lan W, Zhao L*, Su Y*. (2018) PpV, acting via the JNK pathway, represses apoptosis during normal development of Drosophilawing. Apoptosis23: 554-562.
23. Lv Z, Rosenbaum J, Aspelmeier T, and Großhans J*. (2018) A 'molecular guillotine' reveals the interphase function of Kinesin-5. Journal of Cell Science 131(3).
24. Kaiser F, Lv Z, Rodrigues D, Rosenbaum J, Aspelmeier T, Großhans J and Alim K*(2018). Mechanical Model of Nuclei Ordering in DrosophilaEmbryos Reveals Dilution of Stochastic Forces. Biophysical Journal 114(7): 1730-1740.
25. Schmidt A#, Lv Z# and Großhans J* (2018). ELMO and Sponge specify subapical restriction of Canoe and formation of the subapical domain in early Drosophila embryos. Development 145(2).
26. Zhao L, Wang L, Chi C, Lan W, Su Y*. (2017) The emerging roles of phosphatases in Hedgehog pathway. Cell Communication and Signaling 15: 35.
27. Liu M, Li Y, Liu A, Li R, Su Y, Du J, Li C, Zhu AJ. (2016) The exon junction complex regulates the splicing of cell polarity gene dlg1 to control Wingless signaling in development. eLife5: e17200.
28. Du J, Zhang J, He T, Li Y, Su Y, Tie F, Liu M, Harte P, Zhu AJ. (2016) Stuxnet facilitates the degradation of polycomb protein during development. Developmental Cell 37: 507-519.
29. Han P, Bloomekatz J, Ren J, Zhang R, Grinstein JD, Zhao L, Burns CG, Burns CE, Anderson RM, and Chi NC. (2016) Coordinating cardiomyocyte interactions to direct ventricular chamber morphogenesis. Nature 534: 700-704.
30. Lv Z. and Großhans J *(2016). A Radial Actin Network in Apical Constriction. Dev Cell 39(3): 280-282.
31. Jahangiri L, Sharpe M, Novikov N, González-Rosa JM, Borikova A, Nevis K, Paffett-Lugassy N, Zhao L, Adams M, Guner-Ataman B, Burns CE, Burns CG. (2015) The AP-1 transcription factor component Fosl2 potentiates the rate of myocardial differentiation from the zebrafish second heart field. Development 143: 113-122.
32. Mahmoud A, O’Meara C, Gemberling M, Zhao L, Bryant DM, Zheng RM, Gannon JB, Cai L, Choi WY, Egaczyk GF, Burns CG, Burns CE, Macrae CA, Poss KD, Lee RT. (2015) Nerves regulate cardiomyocyte proliferation and heart regeneration. Developmental Cell 34: 387-399.
33. Harrison MR, Bussmann J, Huang Y, Zhao L, Osorio A, Burns CG, Burns CE, Sucov HM, Siekmann AF and Lien CL. (2015) Chemokine guided angiogenesis directs coronary vasculature formation in zebrafish. Developmental Cell 33(4): 442-454.
34. Jang IH, Lu YF, Zhao L, Wenzel PL, Kume T, Datta SM, Arora N, Guiu J, Lagha M, Kim PG, Schlaeger TM, Zon LI, Gigas A, Burns CE, Daley G. (2015) Notch1 acts via Foxc2 to induce the endothelial to hematopoietic transition during embryonic development. Blood 125(9): 1418-1426.
35. Winkler F#, Gummalla M#, Kunneke L#, Lv Z, Zippelius A, Aspelmeier T and Großhans J* (2015). Fluctuation Analysis of Centrosomes Reveals a Cortical Function of Kinesin-1. Biophysical Journal 109(5): 856-868.
36. Zhao L, Borikova AL, Ben-Yair R, Guner-Ataman B, Macrae CA, Lee RT, Burns CG, Burns CE. (2014) Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. PNAS 111(4): 1403-1408.
37. Geisbrecht ER, Sawant K, Su Y, Liu ZC, Silver DL, Burtscher A, Wang X, Zhu AJ, McDonald JA. (2013) Genetic interaction screens identify a role for hedgehog signaling in Drosophila border cell migration. Developmental Dynamics 242: 414-431.
38. Yan S.#, Lv Z#, Winterhoff M#, Wenzl C, Zobel T, Faix J, Bogdan S and Großhans J*. (2013) The F-BAR protein Cip4/Toca-1 antagonizes the formin Diaphanous in membrane stabilization and compartmentalization. Journal of Cell Science (Pt 8): 1796-1805.
39. Zhang J, Liu M, Su Y, Du J, Zhu AJ. (2012) A targeted in vivo RNAi screen reveals deubiquitinases as new regulator of Notch signaling. G3: Genes, Genomes, Genetics 2: 1563-1575.
40. Zheng XF, Yang SY, Han YC, Zhao XY, Zhao L, Tian T, Tong J, Xu P, Xiong C, Meng AM. (2012) Loss of zygotic NUP107 protein causes missing of pharyngeal skeleton and other tissue defects with impaired nuclear pore function in zebrafish embryos. The Journal of Biological Chemistry 287(45): 38254-38264.
41. Du J, Zhang J, Su Y, Liu M, Ospina JK, Zhu AJ. (2011) in vivo RNAi Screen reveals neddylation genes as novel regulators of Hedgehog signaling. PLoS ONE 6: e24168.
42. Su Y#, Ospina JK#, Zhang J#, Michelson A, Schoen AM, Zhu AJ. (2011) Sequential phosphorylation of Smoothened transduces graded Hedgehog signaling. Science Signaling 4: ra43.
43. Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, Guner-Ataman B, Burns CG, Burns CE. (2010) The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis.Development137(11): 1887-1896.
44. Zhao XY, Zhao L, Tian T, Zhang Y, Tong J, Zheng XF, Meng AM. (2010) Interruption of cenph causes mitotic failure and embryonic death, and its haploinsufficiency suppresses cancer in zebrafish.The Journal of Biological Chemistry 285(36): 27924-27934.
45. Tian T, Zhao L, Zhao XY, Zhang M, Meng AM. (2009) A zebrafish gene trap line expresses GFP recapturing expression pattern of foxj1b.Journal of Genetics and Genomics 36(10): 581-589.
46. Tian T#, Zhao L#, Zhang M, Zhao XY, Meng AM. (2009) Both foxj1a and foxj1b are implicated in left-right asymmetric development in zebrafish embryos. Biochemical and Biophysical Research Communications 380(3): 537-542.
47. Zhao L#, Zhao XY#, Tian T#, Lu QL, Nirma Skrbo-Larssen, Wu D, Kuang Z, Zheng XF, Han YC, Yang SY, Zhang CM, Meng AM. (2008) Heart-specific isoform of tropomyosin4 is essential for heartbeat in zebrafish embryos. Cardiovascular Research 80(2): 200-208.
48. Su Y#, Zhang L#, Gao X, Meng F, Wen J, Zhou H, Meng A, Chen Y. (2007) The evolutionally conserved activities of Dapper2 in antagonizing TGF-b signaling. FASEB Journal 21: 682-690.
49. Lin X#, Duan X#, Liang Y#, Su Y#, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen Y, Meng A, Feng X. (2006) PPM1A functions as a Smad phosphatase to terminate TGFb signaling.Cell 125: 915-928.
50. Zhang L#, Zhou H#, Su Y#, Sun Z, Zhang H, Zhang L, Zhang Y, Ning Y, Chen Y, Meng A. (2004) Zebrafish Dpr2 inhibits mesoderm induction by promoting degradation of Nodal receptors. Science306: 114-117.
51. Su Y and Meng A. (2002) The expression of gbx-2 during zebrafish embryogenesis. Mechanisms of Development 113: 107-110.